DeepRIP
m6A peak calling at single-nucleotide-resolution by
deep learning-based algorithm
Identify m6A peak at single-nucleotide-resolution
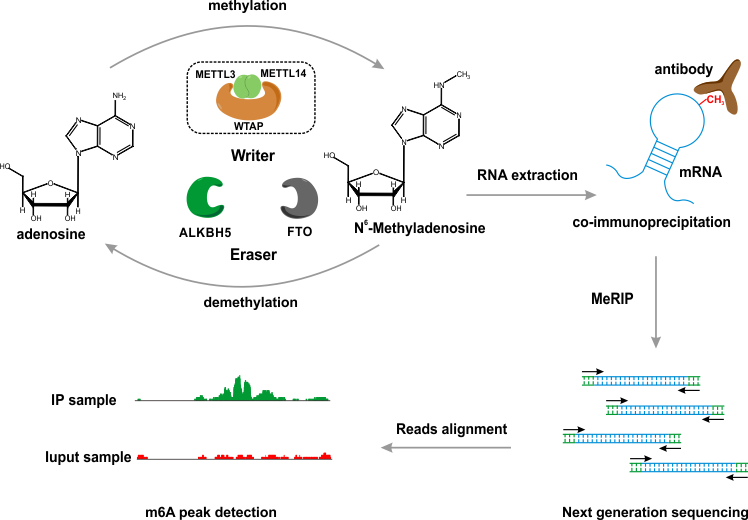
DeepRIP is specifically designed for the detection of m6A peaks from MeRIP-seq experiment at single-nucleotide-resolution.
The application of Restricted Boltzmann Machine achieved a far more accurate derivation of m6A peaks comparing to the state-of-art methods. Additionally, DeepRIP also deploys a Multilayer Perceptron model to identify m6A sites from the detected peaks, enabling a single-nucleotide-resolution peak calling.
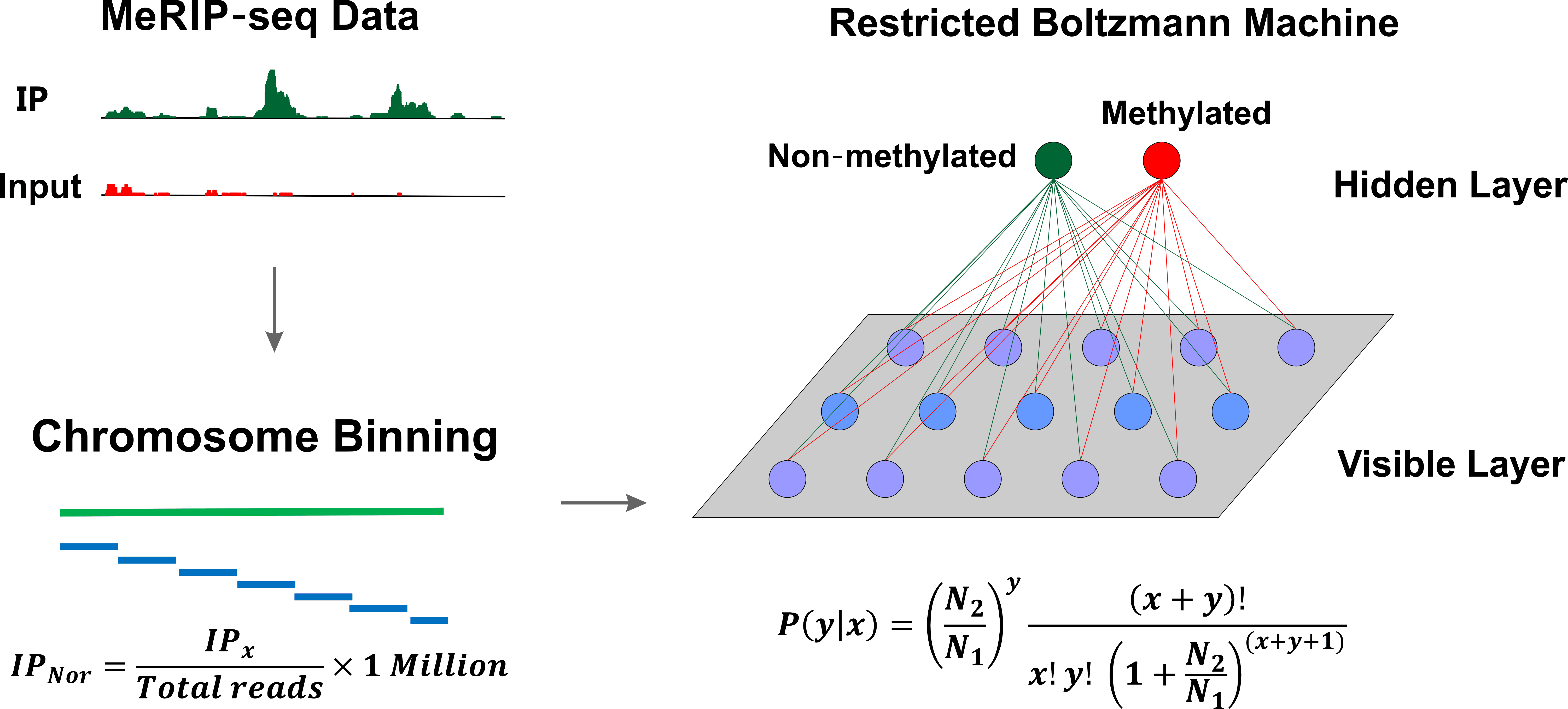
Restricted Boltzmann Machine for peak calling
In order to identify regions that contain m6As, the algorithm first divides the entire genome into a set of mutually connected bins. According to Audic’s model, the chromosomal bins are then parameterized into a set of vectors. For m6A peak calling, the RBM model is built and trained to fit the probability distribution of the input vectors.
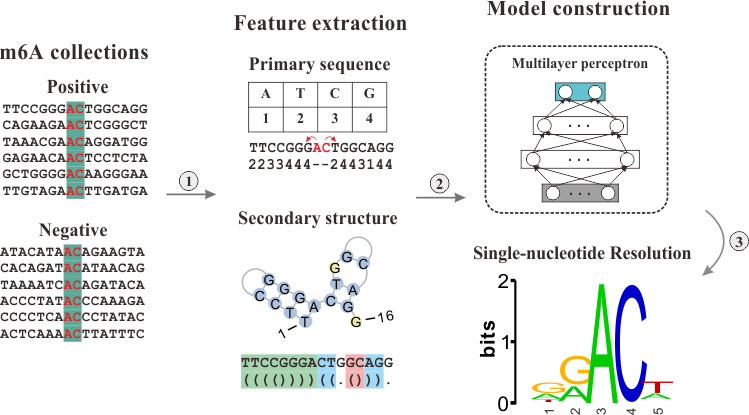
Multilayer Perceptron for m6A sites prediction
To improve the resolution for m6A peak calling, we constructed a multilayer perceptron model to predict precise m6A sites from peak regions. Combining the primary sequence and secondary structure features, a robust prediction model was built. After evaluation, the model is proved to be of accuracy larger than 85%.